About me
In August 2019, I joined the metrology and inspection team at ASML in San Jose, CA.
From 2014 to 2019, I obtained my PhD degree from the Department of Electrical and Computer Engineering of the University of Virginia. I worked on biomedical image processing projects under the guidance of Prof. Daniel Weller. My main research interests include image quality assessment, 3D image reconstruction, and biomedical image segmentation.
From 2010 to 2014, I completed my bachelor’s degree with honor in Information Engineering from Shanghai Jiao Tong University under the guidance of Prof. Weiyao Lin.
Highlighted Projects
1. 3D Mouse Brain Reconstruction and Analysis
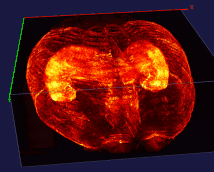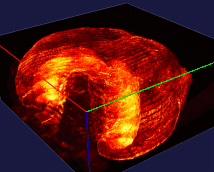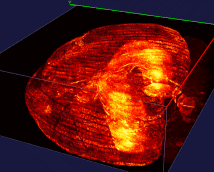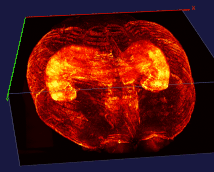
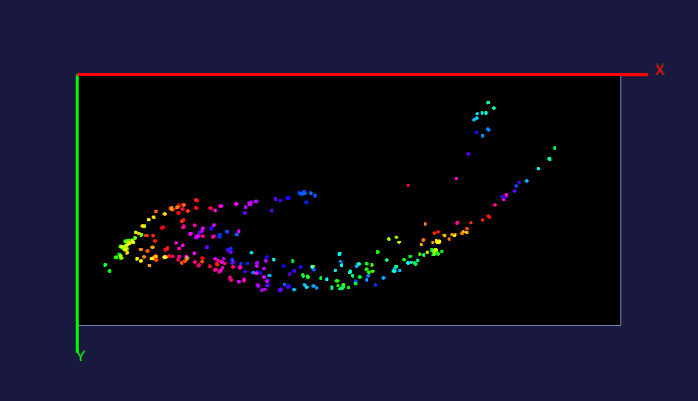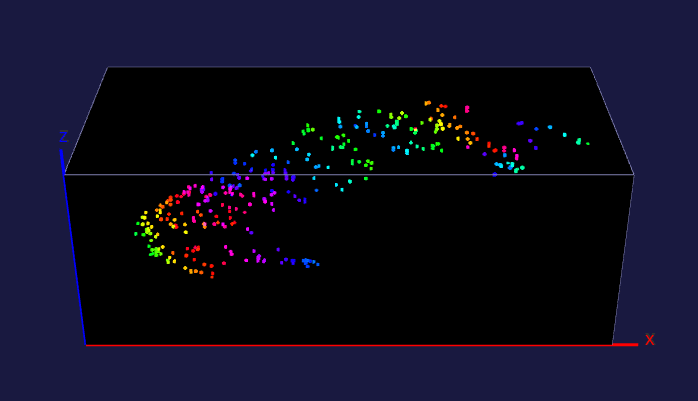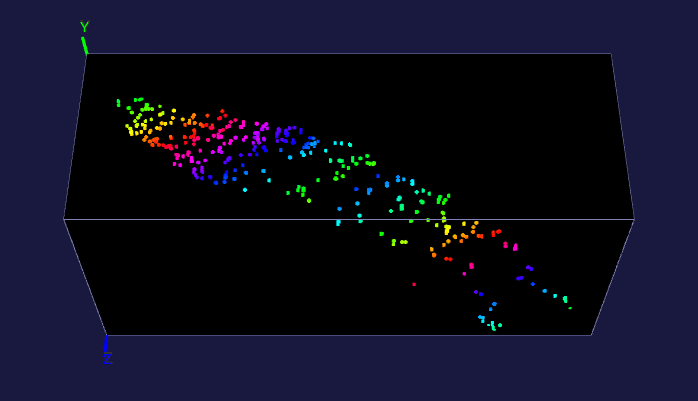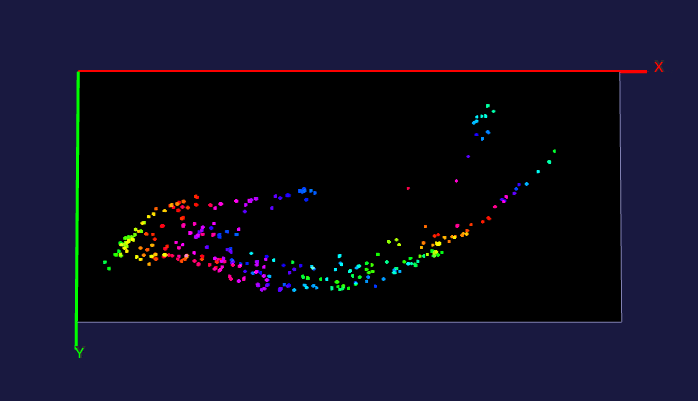
The left image: The high resolution and flexible choices of stains make the microscopy an irreplaceable tool to observe fine brain structures, such as cells. The raw data (~20 brain sections after tissue clearing) of the reconstructed brain on the left is acquired by Dr. Kapur’s lab, and the red fluorescence of tdTomato reflects the seizure spreading during epilepsy. Visualizing and analyzing the seizure spreading pathway help neuroscientists find out the mechanism of epilepsy and the drugs to terminate epilepsy an early stage. Details about the reconstruction method can be found here.
The right image: The dentate gyrus of adult mice contains millions of granule cells, but only a few hundreds to a few thousands of these cells are activated to differentiate environments. The sparse coding by sparse activation in the mouse brain is linked to the task of exploring new environments. This image on the left shows the activated granule cells in the dentate gyrus. More details about this work can be found here.
2. Cell Counting and Segmentation
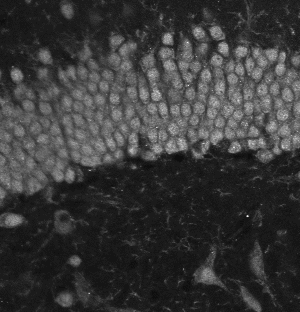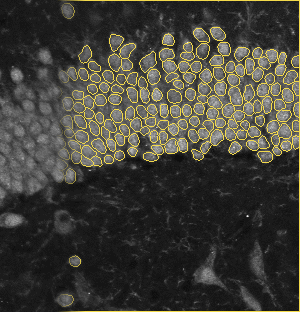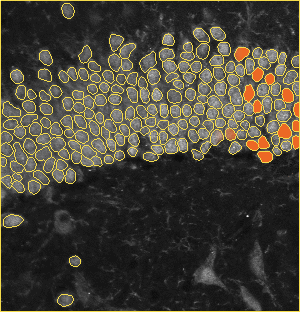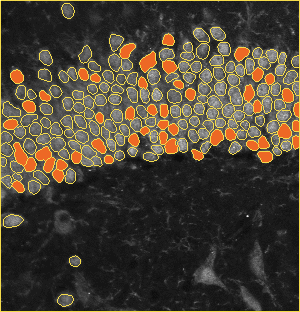
The dentate gyrus is one of the most active regions in the hippocampus. Numerous brain activities are proved to be related to the dentate gyrus, such as the formation of episodic memories and exploration of novel environments. The image on the left is a zoom-in view of the dentate gyrus. The brain specimens in this experiment are double stained by the fluorescence of NeuN and tdTomato. NeuN labels all the nucleus, and tdTomato indicates the activated ones. The segmentation method is based on U-net.
3. Neuron Image Enhancement
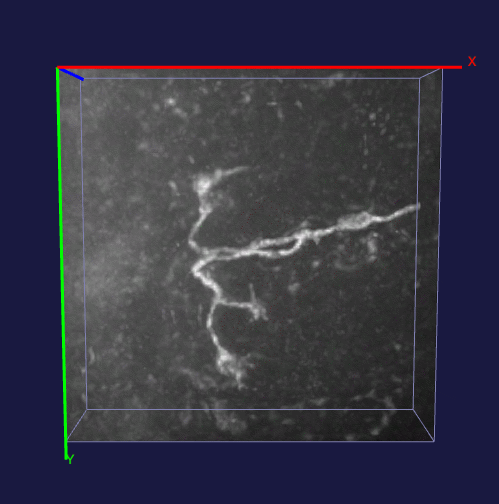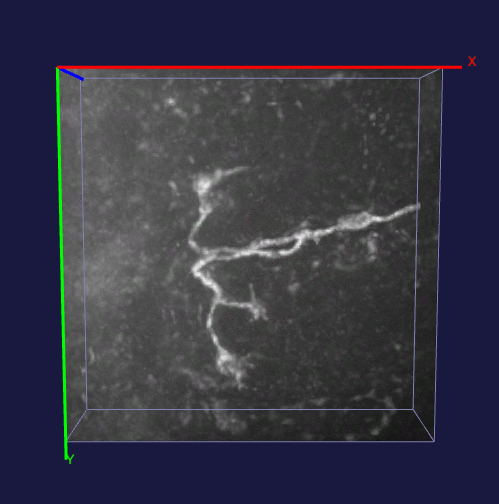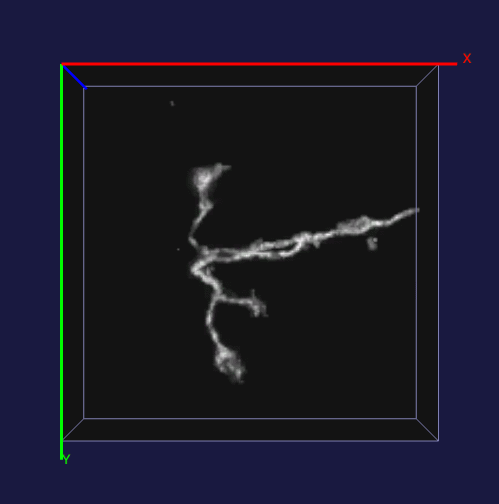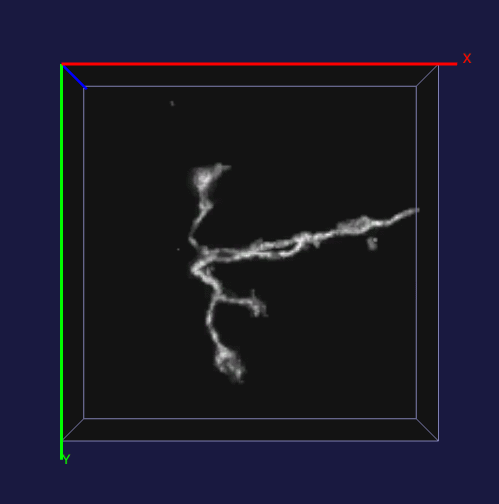 How to effectively capture the image prior information and incorporate them into the solution of a specialized imaging application is critical to achieve state-of-the-art results for many specialized imaging tasks, such as microscopic imaging for biomedical study. In this project, enhancement for images with filamentous structure is studied. Two unique properties for neuron images, the gradient sparsity and the tubular structure are exploited as the key structure priors to achieve the clarity boost for raw neuron images. Paper and code for this work.
How to effectively capture the image prior information and incorporate them into the solution of a specialized imaging application is critical to achieve state-of-the-art results for many specialized imaging tasks, such as microscopic imaging for biomedical study. In this project, enhancement for images with filamentous structure is studied. Two unique properties for neuron images, the gradient sparsity and the tubular structure are exploited as the key structure priors to achieve the clarity boost for raw neuron images. Paper and code for this work.
4. Comparison-based Image Quality Assessment
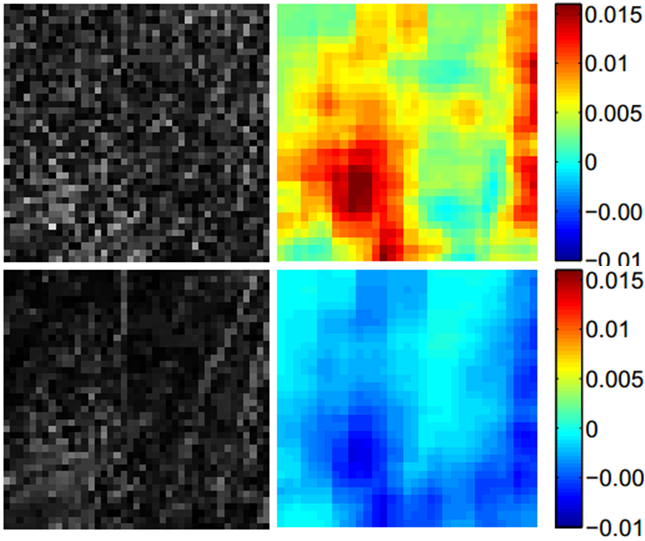
When humans performing image quality assessment, the intuitive approach is through comparision. In the comparison-based IQA proposed in this project, a relative quality index is assigned to the two input images. The implementation of the comparison based approach contains two modules: content detection module and contribution module. The module of content detection determines whether the difference between two input images contains any meaningful structure and the module of contribution is designed to find out which of the two input images mainly contributes to the residual image. The final relative quality index is determined by composing these two modules by the criterion that the input image that contributes to a structured residual is better and the input image that contributes to a random residual is worse.
